:research projects
Identifying factors that regenerate injured hepatocytes using CRISPR screens
We perform CRISPR screens to identify reprogramming factors ameliorating disease-associated transcriptional circuits in human hepatocytes. We apply this technology to models of lipotoxicity and cholestasis in primary human hepatocytes, iPSC-multi-lineage organoids and human ductal organoids. Among different assays, we harness artificial intelligence-guided microscopy that allows label-free, high-throughput analysis of CRISPR screens. Ultimately, we aim to understand single-cell transcriptional circuits and perturbations in human hepatocytes relevant to chronic liver diseases, e.g., non-alcoholic steatohepatitis (NASH). Importantly, we hope to overcome the limitations of mouse and rat studies.
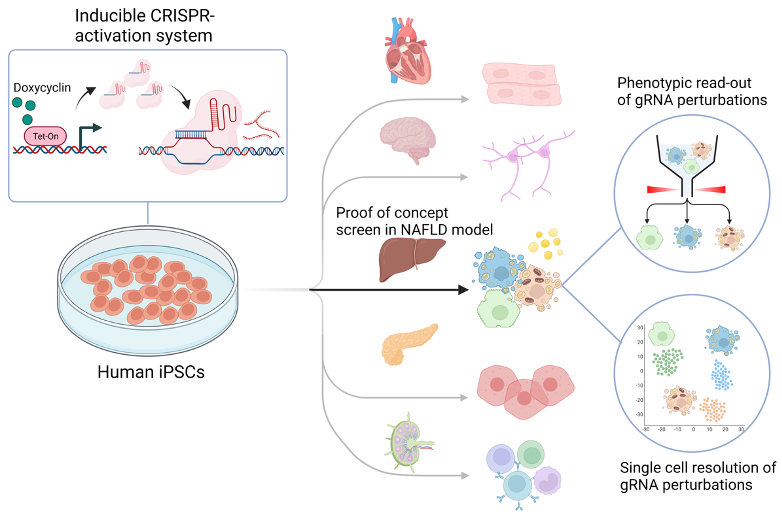
In vivo Liver Reprogramming
Developing regenerative therapies for chronic liver injuries is demanding because little is known about human hepatocyte transcriptional programs providing resilience and regeneration in vivo. We perform in vivo competitive gene selection screens to identify transcription factors and zonal signals restoring function and tissue regeneration in humanized models of chronic liver injury.
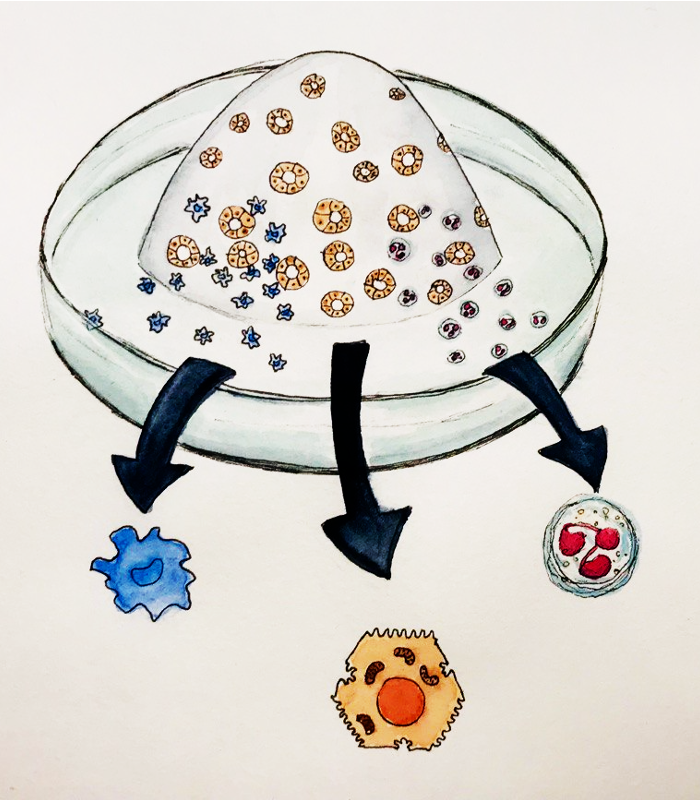
Modeling the human fetal liver with Hepato-hematopoietic Organoids from Pluripotent Stem Cells
The mammalian fetal liver harbors hematopoietic stem and progenitor cells. We lineage-engineer from endoderm and mesoderm human fetal liver organoids that co-develop different syngeneic lineages, e.g., hepatoblasts, stellate cells, hematopoietic progenitor cells and different immune cell lineages, e.g., macrophages and granulocytes. Single-cell multi-omics and bioinformatic analyses help us to understand lineage bifurcations and cell-cell interactions. Ultimately, this human fetal liver organoid platform could serve for the studies of infant leukemias or newborn inflammatory liver diseases.
Organoid-Modeling of biliary autoinflammation
Nearly half of all pediatric patients requiring liver transplantation are caused by Biliary atresia (BA), with a great need for innovation. However, today’s models of BA fail to recapitulate the crosstalk between different lineages, i.e., cholangiocytes and autoreactive immune cell lineages. We perform multi-omics analyses of BA patient liver tissue and apply iPSC-organoid technology to reverse-engineer BA phenotypes with human biliary organoids harboring syngeneic myeloid and lymphoid subsets. Ultimately, we focus on drivers and mechanisms of biliary cytotoxicity.
Collaborators:
Prof. Dr. med. Philip Bufler, Dr. Tilman Breiderhoff (Charité, Pediatric Gastro-Nephro)
Prof. Dr. Christian Conrad (Berlin Institute of Health “Intelligent Imaging”) and Dr. Lorenz Robert Chua
Prof. Dr. med. Frank Tacke, Dr. Adrien Guillot (Charité CVK-Adult Pediatric Gastro-Nephro)
Prof. Takanori Takebe (Cincinnati Children’s Hospital Medical Center, USA; Tokyo Medical and Dental University) and team
Dr. Namshik Han (Milner Therapeutics Institute / Cambridge Centre for AI in Medicine, Department of Applied Mathematics and Theoretical Physics
University of Cambridge)
Prof. Wolfram Goessling (Director, Division of Gastroenterology Massachusetts General Hospital, Harvard Medical School; Director – Harvard–MIT Program of Health Sciences and Technology)
Prof. Dr. Igor Sauer, Dr. Nils Haep, Peter Tang (Charité, Experimentelle Chirurgie)
Dr. Dubravka Vucicevic (Max-Delbrück Center, Ohler Lab)
Dr. Katarzyna Ludwik, Dr. Harald Stachelscheid (BIH Core Unit pluripotent Stem Cells and Organoids (CUSCO)
TRI Thinking Research Instruments GmbH, Hamburg
Prof. Ludovic Vallier (BIH-Charité, Einstein Center; Max Planck Institute for Molecular Genetics, Berlin; Cambridge University/UK)
CellBricks (Organ/Tissue-Bioprinting Company based in Berlin)